Metabox 2.0
Metabox 2.0: A toolbox for thorough metabolomic data analysis, integration and interpretation.
Description
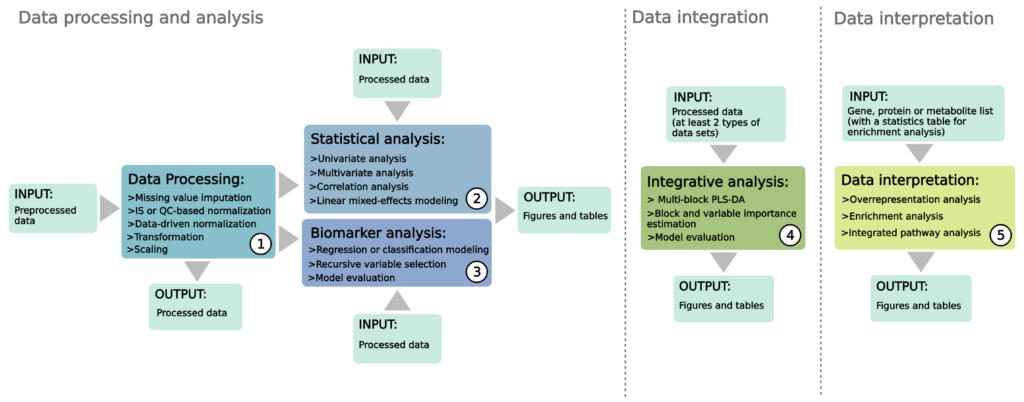
Metabox 2.0 is an updated version of the R package Metabox, released in 2016. The tool includes several methods for data processing, statistical analysis, biomarker analysis, integrative analysis and data interpretation. Metabox 2.0 supports a wide range of users, from bench biologists to experienced bioinformaticians. It comes with an intuitive web interface for simple data analysis. We recommend the R command line version for custom pipelines and other exclusive projects.
How to use
Here are some alternative ways to use Metabox 2.0:
Publications
- Wanichthanarak K, In-On A, Fan S, Fiehn O, Wangwiwatsin A, Khoomrung S. Data processing solutions to render metabolomics more quantitative: case studies in food and clinical metabolomics using Metabox 2.0. Gigascience. 2024 Jan 2;13:giae005. doi: 10.1093/gigascience/giae005.
- Wanichthanarak K, Fan S, Grapov D, Barupal DK, Fiehn O. Metabox: A Toolbox for Metabolomic Data Analysis, Interpretation and Integrative Exploration. PLoS One. 2017 Jan 31;12(1):e0171046. doi: 10.1371/journal.pone.0171046.