CRISP II: Leveraging Deep Learning for Multiclass Phenotyping in GC GC-TOFMS of End-Stage Renal Disease Patients
- General
Information
Two-dimensional gas chromatography time-of-flight mass spectrometry (GC GC-TOFMS) is a highly effective technique for metabolomics analysis, but leveraging its contour images for metabolite profiling is challenging due to the scarcity of metabolite identification libraries and advanced bioinformatics tools. To accelerate the analysis of GC GC-TOFMS data, we developed CRISPII, a deep learning approach that utilizes combination of multiple operations including adaptive gradient thresholding-based identification, simulation, for effective profiling of multiclass separations in GC GC-TOFMS metabolomics data. This study presents substantial improvements over our earlier CRISP software, which was limited to classifying two types of GC GC-TOFMS metabolomic data. CRISP enhances the utility of aggregate feature representative images and introduces multiclass-compatible gradient thresholding-assisted ROI stacking for profiling GC GC-TOFMS contours. The CRISP pipeline supports semi-automatic ROI identification for feature enrichment and classification of GC GC-TOFMS contour images. Additionally, it includes contour synthesis through a generative adversarial network for data augmentation in cases of sparse samples. This manual guides users in utilizing the various options available in CRISP to tailor it to specific datasets or operations. The software full source codes and datasets are be available at GitHub [ https://github.com/vivekmathema/CRISP ]
2. Software Architecture and Operation Guidelines
CRISP is designed to directly input GC GC-TOFMS contour images generated from ChromaTOF for training and profiling. It comprises three main components:
CRISP-II Workflow
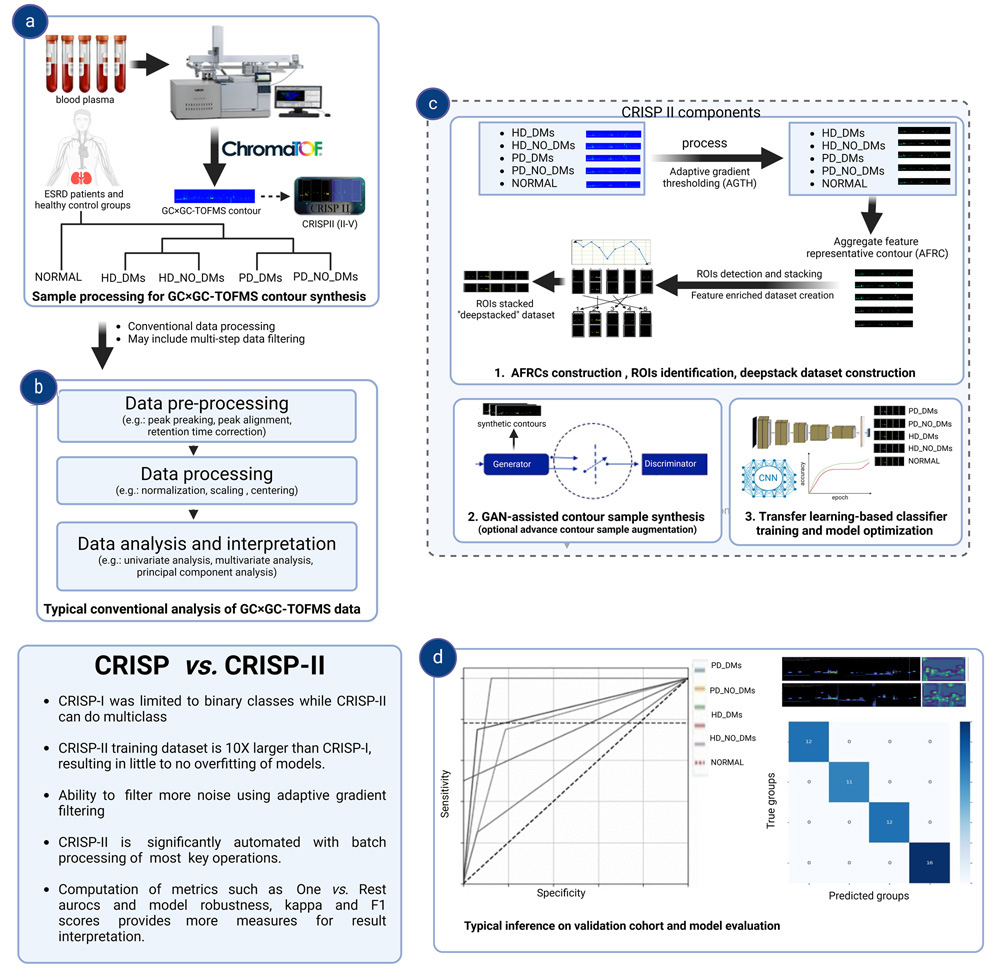
CRISPII provides a user-friendly interface for analyzing GC GC-TOFMS contour images from blood plasma samples of DKD patients and healthy controls. The pipeline supports diverse experimental designs and involves:
(a) Data
Import: Preprocess and import GC GC-TOFMS contour images.
(b) Conventional Analysis: Perform standard metabolomics methods (e.g.,
PCA).
(c) Feature Extraction: Build enriched contour datasets, with optional
GAN-based augmentation and transfer learning to train a multiclass CNN
classifier.
(d) Model Validation and Usage: Validate the trained classifier model
and infer results on unknown samples.
A. ROI & Deep Stacking
i. Manual
extraction of GC GC-TOFMS contour images for pre-processing
ii. Construction of single and multiclass Aggregate Feature Representative
image (AFRC)
iii. Creation of single and multiclass deep stacking datasets
B. GAN-based GC GC-TOFMS Contour Image generator Training & Synthesis
C. Multiclass GC GC-TOFMS Contour classifier training & Inference
Each module can function independently or in combination, depending on the desired outcome. However, this approach can also be adapted for other contour images with similar 2D data structures. Note: All steps from A to C should be performed using the same mode during the inference of an unknown sample.
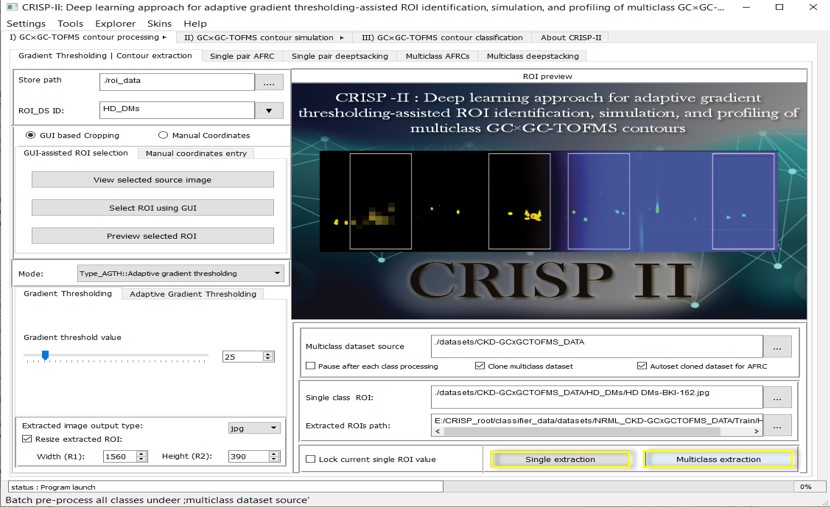
A. ROI & Deep Stacking i. Manual GC GC-TOFMS contour image extraction for pre-processing
This section allows users to manually select a single ROI either graphically or by inputting coordinates. It supports cropping the region to obtain a desired ROI (or image frame in this context) with various options such as resizing, applying image filters, and choosing the output file type. The extraction of initial contour data can be performed in Normal RGB color mode (no color changes), adaptive gradient thresholding (AGTH), or fixed gradient thresholding (FXTH) mode, depending on the user's preference. Once the contour data is extracted in a specific mode, the same mode setting must be used for all contour groups and future inferencing. Selection of an ROI is required for only one image per group folder, and all images in the folder will automatically have the same ROIs extracted in batch operation. For more details, refer to the CRISP-II User Manual.
ii. Single and multiclass Aggregate feature representative contour image construction
This innovative procedure generalizes groups of contour images belonging to a specific class or group through weighted aggregate feature extraction. It enables dominant features to be prominently represented in the final single representative image, while rare or outlier features are minimized algorithmically. This process yields an aggregate feature representative contour (AFRC) image that represents the common dominant features within the group, reducing manual selection bias. Parameters such as FPS, weight accumulation factor, and cyclic image duration control the number of features included in the final AFRC image. Various image augmentation techniques (e.g., blur, noise, erosion, dilation) can be applied during AFRC construction to enhance feature coverage. The AFRC image can then be automatically utilized in the subsequent module for Auto ROI detection and Deepstacking. This operation can also be performed in batches for all groups. For more details, refer to the CRISP-II User Manual.
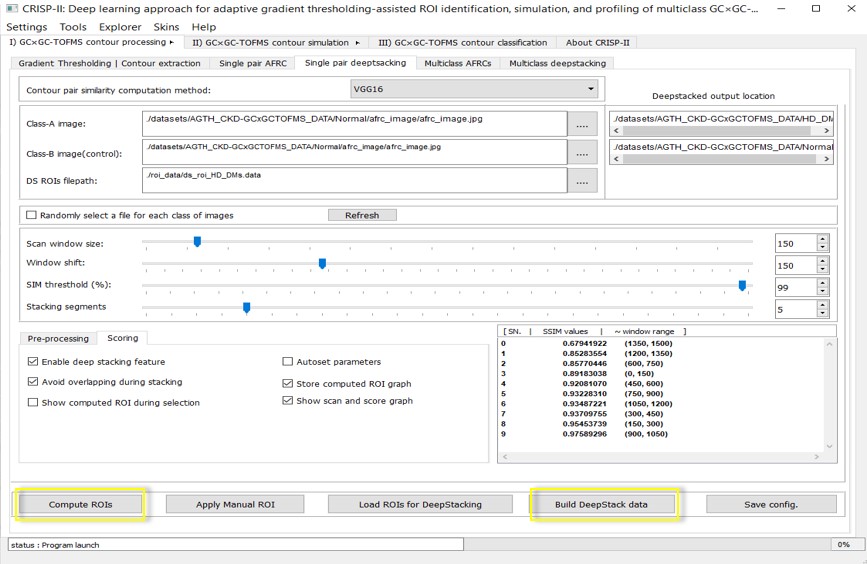
iii. ROIs identification and Single/Multiclass deepstacking dataset construction
This module performs several critical tasks for building the final classifier training database for contour simulation and classification. It includes various customized models and algorithms to identify multiple ROIs between two AFRC images, such as retrained VGG16 (default), SIAMESE, SSIM, PNSR, Hamming, and FID (details in the publications). The top-N (N=5 by default) ROIs are identified based on user-defined settings like similarity threshold, scanning window size, and window overlap amounts. During processing, the scanning window slider evaluates contour data features in AFRC images of each class (e.g., Disease class vs. Control) from left to right, generating scores for each window area and a list of coordinates with scores for the top-N (default N =5) least similar regions. Users can view a graph of these scores along the R1 dimension of the contour data. Subsequently, contour feature images for all individual contours in each group can be generated as "DeepStacked" images using the DeepStack dataset builder. This feature-stacked dataset can be utilized for simulation to amplify contrasting features between different profiles. Once the DeepStacking dataset is created, it can simulate feature vectors for further classification. This operation can also be performed pairwise (single pair Deepstacking tab) or in batch mode for multiclass data (multiclass Deepstacking tab). For more details, refer to the CRISP-II User Manual.
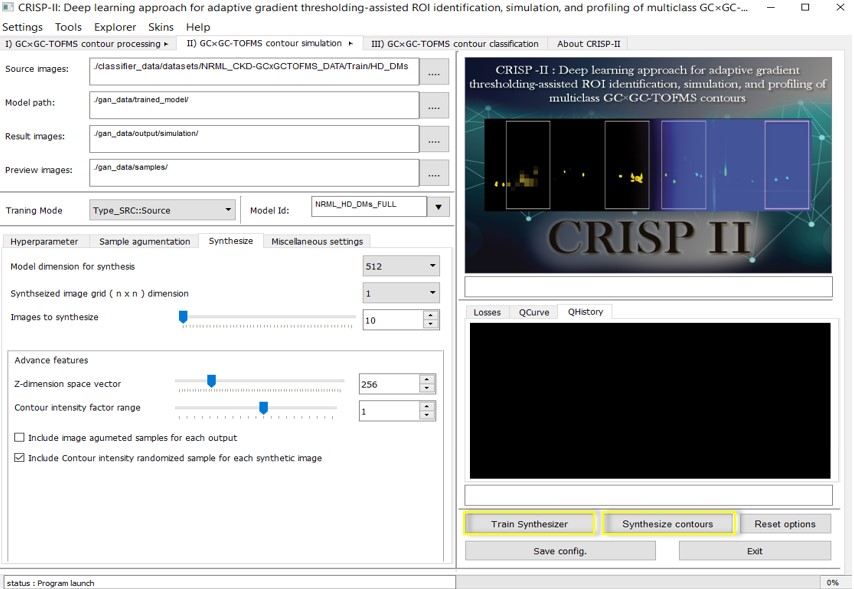
B. GAN-based GC GC-TOFMS contour image training & synthesis**
CRISP incorporates advanced contour simulation neural networks based on Generative Adversarial Networks (GANs), offering extensive customization options. The generator can train models using contour images of 512 512 pixels, even with a small sample size, and still achieve good results. Larger sample sizes lead to improved outcomes due to the greater diversity of the original images. Users can fine-tune hyperparameters and other settings such as FIDs, Z-vectors, RELU, learning rates, and model optimizers (II GC GC-TOFMS Tab -> Hyperparameters).
In the image augmentation tab (Contour Synthesis -> Augmentation), users can apply various filters (e.g., dilation, distortion, erosion, contrast, brightness, noise, edge sharpening) to initially increase the diversity of limited contours fed into the generator, enhancing the dataset without significantly altering the overall profile of the input contours. This is particularly advantageous in low-sample conditions. Real-time graphs of model loss and previews, along with FID curves, are displayed during training to monitor the generator's progress. The qScore (forming the qCurve) is a custom metric that indicates the quality of the output (0.0000-1.8), primarily based on the sharpness or blurriness of the synthetic image compared to the original contour image distribution. If the sharpness of images closely matches the source data, the qScore will be around ~1.8. Although not entirely reliable, it provides a trend for ongoing simulation training as a real-time feature update.
Training models and their history can be saved or restored at any point during the training, along with the configuration. Once the model is sufficiently trained (based on FID curves, model loss, and preview images), it can be used to synthesize entirely new contour plots without needing the original datasets (B. Contour Synthesis -> 3. Synthesize) in a customizable grid ranging from 1 1 to 10 10. An advanced option allows users to control the intensity of entities in the synthesized contour, which often corresponds to the concentration of metabolites. This feature works best when the source images have minimal background noise or column bleeding. Image augmentation can also be applied to the synthesized output contours, further increasing the diversity of the generated samples. Manipulating the Z-vector can lead to greater diversity in the generated samples. There are numerous other options available for image manipulation and preview modes based on custom requirements. For more details on working protocol, refer to the CRISP-II User Manual.
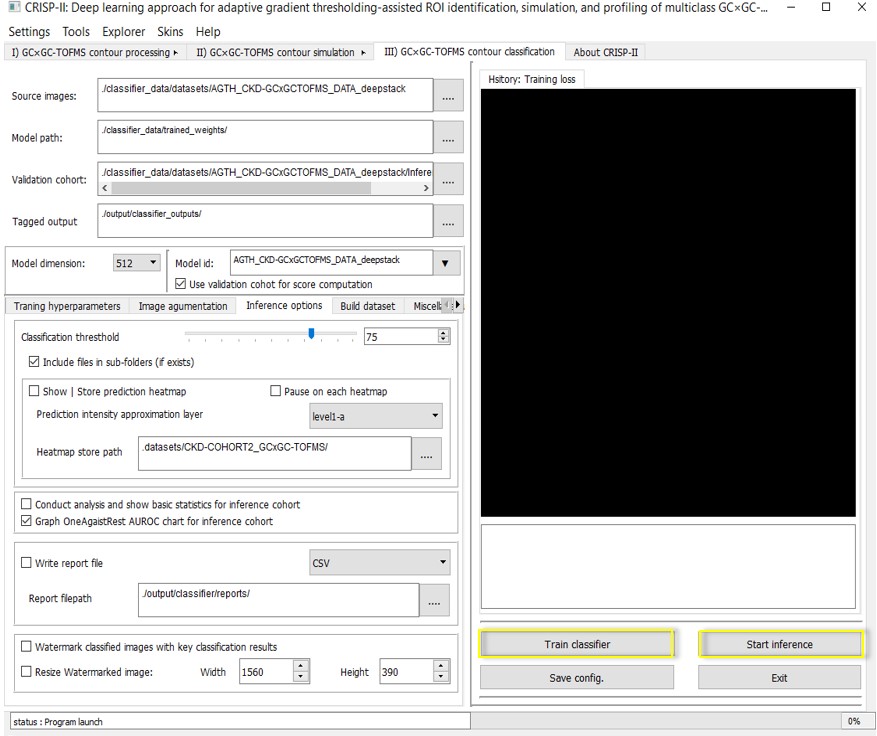
C. Multiclass GC GC-TOFMS contour training & inference**
This module is responsible for training and profiling of GC GC-MSTOF contours with option to output inference result-tagged results and general statistics of test samples.
GC GC-TOFMS
Training:
This is the final module of Contour Profiler, employing customized
state-of-the-art deep convolutional neural networks (D-CNN) like VGG16
(default), VGG19, Inception V3, DenseNet, and ResNet for contour classification using transfer learning.
This approach is effective even with a relatively small sample size. The module
consists of two submodules:
i GC GC-TOFMS Classifier Training: This submodule can train on multiple classes of GC GC-MSTOF data. The datasets are split into training and validation sets to assess training and validation accuracy, receiver operating characteristic (ROC) curves, and model loss functions. It includes a built-in image augmentation option that can randomly apply various image augmentation techniques (e.g., shearing, skewing, and distortion) to increase the diversity of the training data. CRISP also offers the option to view classification performance for a validation cohort if the cohort is provided in the same format as the training/testing set. It also provides one-vs-rest AUROC for evaluating model performance on each class in a multiclass dataset.
ii GC GC-TOFMS
Inference:
This is the final step of the CRISP platform, where the trained classifier
model is used to infer unknown samples based on preset thresholds. The GUI
offers real-time visualization of classification and validation accuracy, as
well as the model loss function, allowing users to instantly review the trained
model's history. The inference step also generates general statistics for the
validation cohort. Heatmaps for the inference samples
can be created along with the watermarked inference output. Models can continue
training with updated datasets, and the training configuration can be saved for
future use and inference on unknown samples. A report file is generated,
including the source images used for inference, and optionally tagged images, heatmaps, and their corresponding classification
confidence. Further details of both training and inference can be found in user
manual.
3. Setup and miscellaneous
Current issues with docker The CRISP-II has major GUI activities which currently is not fully supported causing it failure in UI-associated operation. We are actively looking to this issue and will try to resolve in future. Thus, at present, users can directly utilize the source code and supplied pre-installed environment. Most UI target users that operate ChromaTOF can easily make full use of CRISPII in windows using the provided portable out-of-box package.
Dataset class(or group) name annotation rules --> Classes are represented by their folder name inside main source folder (e.g: HD_DMs, HD_NO_DMs, PD_DMs, PD_NO_DMs,and NORMAL) which is automatically used as class ID --> The folder names should be same for both training & Validation cohort to make results reproducible & correct order of classes during training. --> Model stores the names of classes for annotation of inference results --> All samples for inference, validation or test images should be pre-processed in same order as the classifier training contour image dataset.
Requirements for
Python3 installation
Install the requirement for the minimum GPU version of the python pip install -r requirements_gpu.txt
Install the requirement for the minimum CPU version of the python pip install -r requirements_cpu.txt
NOTE: Current review version of as
passcode protected as supplied in manuscript to maintain data confidentiality.
The standalone windows package for GPU version of CRISPII. This is a
recommended setup for non-technical users. User can directly downloaded the
pre-built package and run the CRISP out-of-the-box
Link:
https://drive.google.com/file/d/1UHxZd10cWxMEIkYmYZM41NIAP91jT8M9/view?usp=sharing
MD5 checksum:
c5c523b20b48622c01f2cc79fae08126
The standalone windows package for CPU version of CRISPII
(Very Slow for training. Slow but relatively simple to install and inference
than GPU version. Will be uploaded upon acceptance of manuscript)
Link: https://drive.google.com/file/d/1hGafZGsIiq7V4Yiqh_kwEGsU0Aivzjqf/view?usp=sharing
MD5 checksum: 6b7bc869ab4d5b4cee0b843de43980bf
The latest updated datasets for
classifier under "./classifier/datasets/trained_weights"
is available as:
Link:
https://drive.google.com/file/d/19gGKyn-D1dVUQW2H3bZUIGib0zv6uXeb/view?usp=sharing
MD5 checksum:
fee766d5141f24cc89152ade824d3fac
For creating Anaconda environment as OS independent version of CRISP-II, we currently recommend to use only CPU version. The GPU version requires moderate to advance CUDA installation knowledge for Linux and may not be suitable for starters.
For GPU Version of Anaconda environment of CRISP. Users may have to tweak the installation versions if any repository needs were updated or conda channel changes. These commands will simply install conda version of the pip3 requirements to run CRISP. May not be suitable for some version of Ubuntu OS.
1) conda create --name GCxGC_CRISPII python=3.6
2.1) conda install --file requirements_cpu.txt (for CPU version, recommended for Linux Ubuntu)
2.1) conda install --file requirements_gpu.txt (for GPU version, recommended for Windows OS)
3) conda activate GCxGC_CRISPII
4) (GCxGC_CRISPII env) conda > python3 crsip.py
CRISP Command line parameters information
python3 CRISP.py [-h | --help]
[--gui_type GUI_TYPE] [--config_run CONFIG_RUN]
[--config_fpath CONFIG_FPATH] [--run_session RUN_SESSION]
Optional arguments:
-h, --help Shows this command line help message and exits CRISP
--gui_type GUI_TYPE
Use different GUI Schemes. Five types available [0: Breeze, 1: Oxygen, 2: QtCurve, 3: Windows, 4:Fusion ]
--config_run CONFIG_RUN [Set 0: False, 1: True]. Run CRISPII in GUI mode ONLY. No configuration modules will be run and CRISP will open GUI with default settings
--config_fpath CONFIG_FPATH full pathname of the configuration file to run. The Confiruation file will be run without any user input or confirmation
--run_session RUN_SESSION [None,gan_train, gan_syn, cls_train, cls_inf] | None : Only loads gui with selected configuration. Following modes are available
gan_train : Load and run gui for GAN model training
gan_syn : Load and run gui for GAN synthesis
cls_train : Load and run gui for classifier training
cls_inf : Load and run gui for classifier inferencing
NOTE: Commandline configuration run is not currently available for ROIs and Deepstacking.
Due to large numbers of parameters the definition of each parameter is commented in configuration file itself.
The
Definitions of most parameters are presented as tool tip text in status bar of
GUI interface.
The CRISP software
will undergo continuous development, with minor bugs being fixed over time. The
primary goal of making the software open source is to enable a larger community
to participate, contribute, and assist in the customization and development of
deep learning-based techniques for GC GC-TOFMS contour image metabolomics.
Website Content
License
Content on this website is licensed
under Creative Commons Attribution 4.0 International (CC BY 4.0). Source code
is available under the MIT License.